en
names in breadcrumbs

Abstract
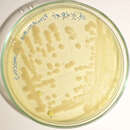
Global emergence of Pseudomonas fluorescens (P. fluorescens) displays a mechanism of resistance to all existing antimicrobials. Due to its strong ability to acquire resistance, there is a need of some alternative treatment strategy. Objective of this study was to investigate the effect of biofield treatment on antimicrobial sensitivity pattern of P. fluorescens. P. fluorescens cells were procured from MicroBioLogics in sealed packs bearing the American Type Culture Collection (ATCC 49838) number. Two sets of ATCC samples were taken in this experiment and denoted as A and B. ATCC-A sample was revived and divided into two groups (Gr) i.e. Gr.I (control) and Gr.II (revived); likewise, ATCC-B was labeled as Gr.III (lyophilized). Gr.II and III were given biofield treatment and were measured by MicroScan Walk-Away® system before and after treatment. Parameters studied in experiment were antimicrobial sensitivity, minimum inhibitory concentration (MIC), biochemical reactions, and biotype number of both control and treatment groups using MicroScan Walk-Away® system. Experimental results showed antimicrobials such as cefepime, cefotaxime, ceftazidime, ceftriaxone, ciprofloxacin, piperacillin, tetracycline, and tobramycin showed altered sensitivity and MIC values in treated group as compared to control. Biochemical reactions showed positive reaction in malonate, melibiose, nitrate, galactosidase, ornithine, raffinose, sorbitol, sucrose, tobramycin and Voges-Proskauer in Gr.II. Arabinose, colistin, glucose, and rhaminose also showed positive reactions in Gr.II on day 10 while arginine and cetrimide showed negative reaction in Gr.III as compared to control. Biochemical tests results revealed a change in biotype number in Gr.II (34101173, day 5), (77103177, a very rare biotype on day 10) and Gr.III (40000043) as compared to control (02041722). Organism was identified as Enterobacter cloacae (GrII, day 10) and Vibrio fluvialis (Gr.III, day 10) with respect to control. These findings suggest that biofield treatment made significant alteration in sensitivity pattern, MIC values, and biotype number of P. fluorescens.Biofield therapies have been reported to improve the quality of life as compared to other energy medicine. The aim of the study was to evaluate the impact of Mr. Trivedi’s biofield energy treatment on Pseudomonas fluorescens (P. fluorescens) for antimicrobial sensitivity, minimum inhibitory concentration (MIC), biochemical reactions, and biotype number. P. fluorescens cells were procured from MicroBioLogics Inc., USA in sealed packs bearing the American Type Culture Collection (ATCC 49838) number and divided in control and treated group. The effect was evaluated on day 10, and 159 after biofield treatment in lyophilized state. Further study was performed on day 5, 10, and 15 after retreatment on day 159 in revived state as per study design. All experimental parameters were studied using automated MicroScan Walk-Away® system. The 16S rDNA sequencing was carried out to correlate the phylogenetic relationship of P. fluorescens with other bacterial species after treatment. The results showed improved sensitivities and decreased MIC value of aztreonam, cefepime, moxifloxacin, and tetracycline in revived and lyophilized treated sample with respect to the control. Arginine, cetrimide, kanamycin, and glucose showed altered biochemical reactions after biofield treatment with respect to control. Biotype numbers were altered along with species in lyophilized as well as in revived group. Based on nucleotides homology and phylogenetic analysis using 16S rDNA gene sequencing, treated sample was detected to be Pseudomonas entomophila (GenBank Accession Number: AY907566) with 96% identity of gene sequencing data, which was nearest homolog species to P. fluorescens (Accession No. EF672049). These findings suggest that Mr. Trivedi’s unique biofield treatment has the capability to alter changes in pathogenic P. fluorescens even in the lyophilized storage condition and can be used to modify the sensitivity of microbes against antimicrobials.
Biofield therapies have been reported to improve the quality of life as compared to other energy medicine. The aim of the study was to evaluate the impact of Mr. Trivedi’s biofield energy treatment on Pseudomonas fluorescens (P. fluorescens) for antimicrobial sensitivity, minimum inhibitory concentration (MIC), biochemical reactions, and biotype number. P. fluorescens cells were procured from MicroBioLogics Inc., USA in sealed packs bearing the American Type Culture Collection (ATCC 49838) number and divided in control and treated group. The effect was evaluated on day 10, and 159 after biofield treatment in lyophilized state. Further study was performed on day 5, 10, and 15 after retreatment on day 159 in revived state as per study design. All experimental parameters were studied using automated MicroScan Walk-Away® system. The 16S rDNA sequencing was carried out to correlate the phylogenetic relationship of P. fluorescens with other bacterial species after treatment. The results showed improved sensitivities and decreased MIC value of aztreonam, cefepime, moxifloxacin, and tetracycline in revived and lyophilized treated sample with respect to the control. Arginine, cetrimide, kanamycin, and glucose showed altered biochemical reactions after biofield treatment with respect to control. Biotype numbers were altered along with species in lyophilized as well as in revived group. Based on nucleotides homology and phylogenetic analysis using 16S rDNA gene sequencing, treated sample was detected to be Pseudomonas entomophila (GenBank Accession Number: AY907566) with 96% identity of gene sequencing data, which was nearest homolog species to P. fluorescens (Accession No. EF672049). These findings suggest that Mr. Trivedi’s unique biofield treatment has the capability to alter changes in pathogenic P. fluorescens even in the lyophilized storage condition and can be used to modify the sensitivity of microbes against antimicrobials.
Pseudomonas fluorescens is a common Gram-negative, rod-shaped bacterium.[1] It belongs to the Pseudomonas genus; 16S rRNA analysis as well as phylogenomic analysis has placed P. fluorescens in the P. fluorescens group within the genus,[2][3] to which it lends its name.
Pseudomonas fluorescens has multiple flagella. It has an extremely versatile metabolism, and can be found in the soil and in water. It is an obligate aerobe, but certain strains are capable of using nitrate instead of oxygen as a final electron acceptor during cellular respiration.
Optimal temperatures for growth of P. fluorescens are 25–30°C. It tests positive for the oxidase test. It is also a nonsaccharolytic bacterial species.
Heat-stable lipases and proteases are produced by P. fluorescens and other similar pseudomonads.[4] These enzymes cause milk to spoil, by causing bitterness, casein breakdown, and ropiness due to production of slime and coagulation of proteins.[5][6]
The word Pseudomonas means false unit, being derived from the Greek words pseudēs (Greek: ψευδής – false) and monas (Latin: monas, from Greek: μονάς – a single unit). The word was used early in the history of microbiology to refer to germs. The specific name fluorescens refers to the microbe's secretion of a soluble fluorescent pigment called pyoverdin, which is a type of siderophore.[7]
Notable P. fluorescens strains SBW25,[8] Pf-5[9] and PfO-1[10] have been sequenced, among others.
A comparative genomic study (in 2020) analyzed 494 complete genomes from the entire Pseudomonas genus, with 25 of them being annotated as P. fluorescens.[3] The phylogenomic analysis clearly showed that the 25 strains annotated as P. fluorescens did not form a monophyletic group.[3] In addition, their Average Nucleotide Identities did not fulfil the criteria of a species, since they were very diverse. It was concluded that P. fluorescens is not a species in the strict sense, but should be considered as a wider evolutionary group, or a species complex, that includes within it other species too.[3] This finding is in accordance with previous analyses of 107 Pseudomonas species, using four core 'housekeeping' genes, that consider P. fluorescens as a relaxed species complex.[11]
The P. fluorescens relaxed evolutionary group that was defined in,[3] on the basis of the genus phylogenomic tree, comprised 96 genomes and displayed high levels of phylogenetic heterogeneity. It comprised many species, such as Pseudomonas corrugata, Pseudomonas brassicacearum, Pseudomonas frederiksbergensis, Pseudomonas mandelii, Pseudomonas kribbensis, Pseudomonas koreensis, Pseudomonas mucidolens, Pseudomonas veronii, Pseudomonas antarctica, Pseudomonas azotoformans, Pseudomonas trivialis, Pseudomonas lurida, Pseudomonas azotoformans, Pseudomonas poae, Pseudomonas libanensis, Pseudomonas synxantha, and Pseudomonas orientalis. The core proteome of the P. fluorescens group comprised 1396 proteins. The protein count and GC content of the strains of the P. fluorescens group ranged between 4152 and 6678 (average: 5603) and between 58.7–62% (average: 60.3%), respectively. Another comparative genomic analysis of 71 P. fluorescens genomes identified eight major subgroups and developed a set of nine genes as markers for classification within this lineage.[12]
There are two strains of Pseudomonas fluorescens associated with Dictyostelium discoideum. One strain serves as a food source and the other strain does not. The main genetic difference between these two strains is a mutation of the global activator gene called gacA. This gene plays a key role in gene regulation; when this gene is mutated in the nonfood bacterial strain, it is transformed into a food bacterial strain.[13]
Some P. fluorescens strains (CHA0 or Pf-5, for example) present biocontrol properties, protecting the roots of some plant species against parasitic fungi such as Fusarium or the oomycete Pythium, as well as some phytophagous nematodes.[14]
It is not clear exactly how the plant growth-promoting properties of P. fluorescens are achieved; theories include:
To be specific, certain P. fluorescens isolates produce the secondary metabolite 2,4-diacetylphloroglucinol (2,4-DAPG), the compound found to be responsible for antiphytopathogenic and biocontrol properties in these strains.[15] The phl gene cluster encodes factors for 2,4-DAPG biosynthesis, regulation, export, and degradation. Eight genes, phlHGFACBDE, are annotated in this cluster and conserved organizationally in 2,4-DAPG-producing strains of P. fluorescens. Of these genes, phlD encodes a type III polyketide synthase, representing the key biosynthetic factor for 2,4-DAPG production. PhlD shows similarity to plant chalcone synthases and has been theorized to originate from horizontal gene transfer.[15] Phylogenetic and genomic analysis, though, has revealed that the entire phl gene cluster is ancestral to P. fluorescens, many strains have lost the capacity, and it exists on different genomic regions among strains.[16]
Some experimental evidence supports all of these theories, in certain conditions; a good review of the topic is written by Haas and Defago.[17]
Several strains of P. fluorescens, such as Pf-5 and JL3985, have developed a natural resistance to ampicillin and streptomycin.[18] These antibiotics are regularly used in biological research as a selective pressure tool to promote plasmid expression.
The strain referred to as Pf-CL145A has proved itself a promising solution for the control of invasive zebra mussels and quagga mussels (Dreissena). This bacterial strain is an environmental isolate capable of killing>90% of these mussels by intoxication (i.e., not infection), as a result of natural product(s) associated with their cell walls, and with dead Pf-145A cells killing the mussels equally as well as live cells.[19] Following ingestion of the bacterial cells mussel death occurs following lysis and necrosis of the digestive gland and sloughing of stomach epithelium.[20] Research to date indicates very high specificity to zebra and quagga mussels, with low risk of nontarget impact.[21] Pf-CL145A has now been commercialized under the product name Zequanox, with dead bacterial cells as its active ingredient.
Recent results showed the production of the phytohormone cytokinin by P. fluorescens strain G20-18 to be critical for its biocontrol activity by activating plant resistance.[22]
By culturing P. fluorescens, mupirocin (an antibiotic) can be produced, which has been found to be useful in treating skin, ear, and eye disorders.[23] Mupirocin free acid and its salts and esters are agents currently used in creams, ointments, and sprays as a treatment of methicillin-resistant Staphylococcus aureus infection.
Pseudomonas fluorescens demonstrates hemolytic activity, and as a result, has been known to infect blood transfusions.[24]
Pseudomonas fluorescens produces the antibiotic Obafluorin.[25][26]
Pseudomonas fluorescens is an unusual cause of disease in humans, and usually affects patients with compromised immune systems (e.g., patients on cancer treatment). From 2004 to 2006, an outbreak of P. fluorescens in the United States involved 80 patients in six states. The source of the infection was contaminated heparinized saline flushes being used with cancer patients.[27]
Pseudomonas fluorescens is also a known cause of fin rot in fish.
Pseudomonas fluorescens produces phenazine, phenazine carboxylic acid,[28] 2,4-diacetylphloroglucinol[29] and the MRSA-active antibiotic mupirocin.[30]
4-Hydroxyacetophenone monooxygenase is an enzyme found in P. fluorescens that transforms piceol, NADPH, H+, and O2 into 4-hydroxyphenyl acetate, NADP+, and H2O.
Pseudomonas fluorescens is a common Gram-negative, rod-shaped bacterium. It belongs to the Pseudomonas genus; 16S rRNA analysis as well as phylogenomic analysis has placed P. fluorescens in the P. fluorescens group within the genus, to which it lends its name.