-
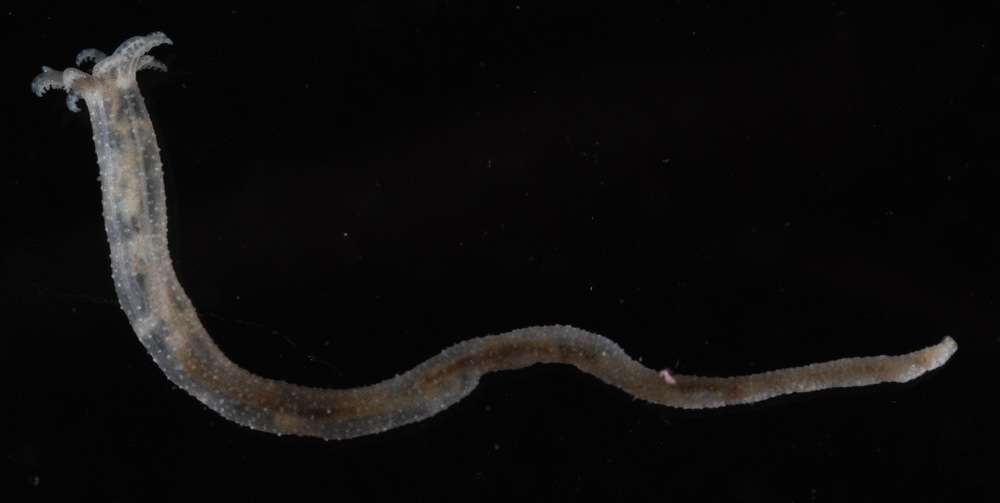
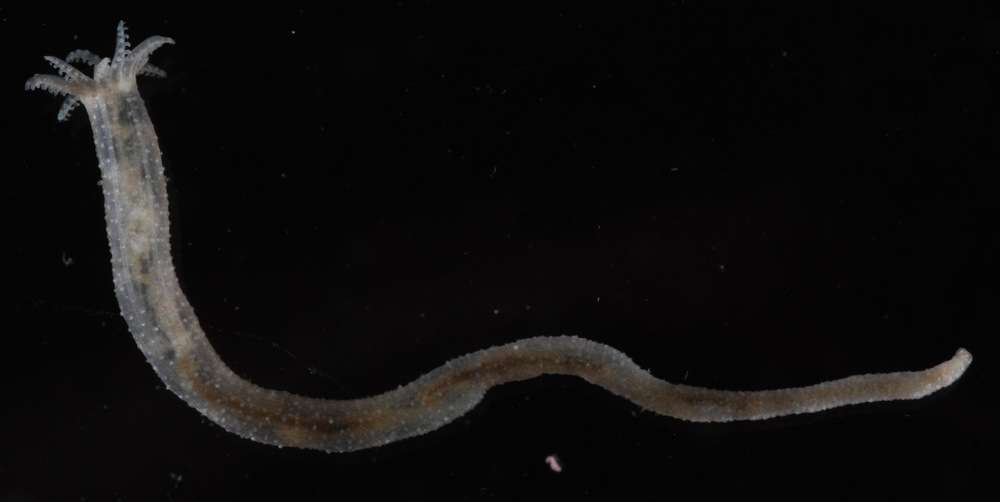
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
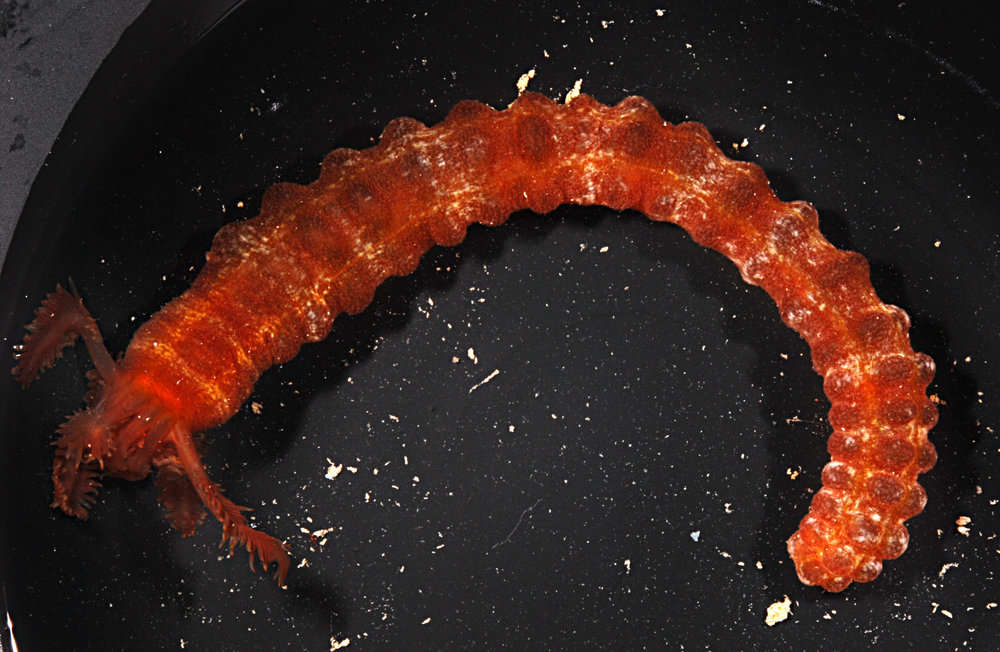
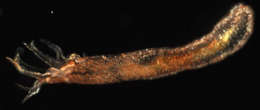
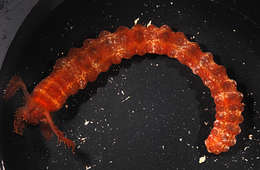
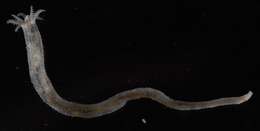
Item Type: Image Title: Chiritid sea cucumber Copyright: Serpent Project Species: Chiridota heheva Behaviour: Moving on Seabed Site: Atlantic -- Gulf of Mexico -- Tiger Site Description: Seafloor Depth (m): 2750 Latitude: 26 deg 10' 48" N Longitude: 94 deg 37' 23" W Countries: Mexico -- Gulf of Mexico Habitat: Temperate Gravel and Sand Rig: Discoverer Deep Seas Project Partners: Transocean, Subsea 7, Chevron USA ROV: Hercules 8 Deposited By: Rob Curry Deposited On: 13 March 2007
-
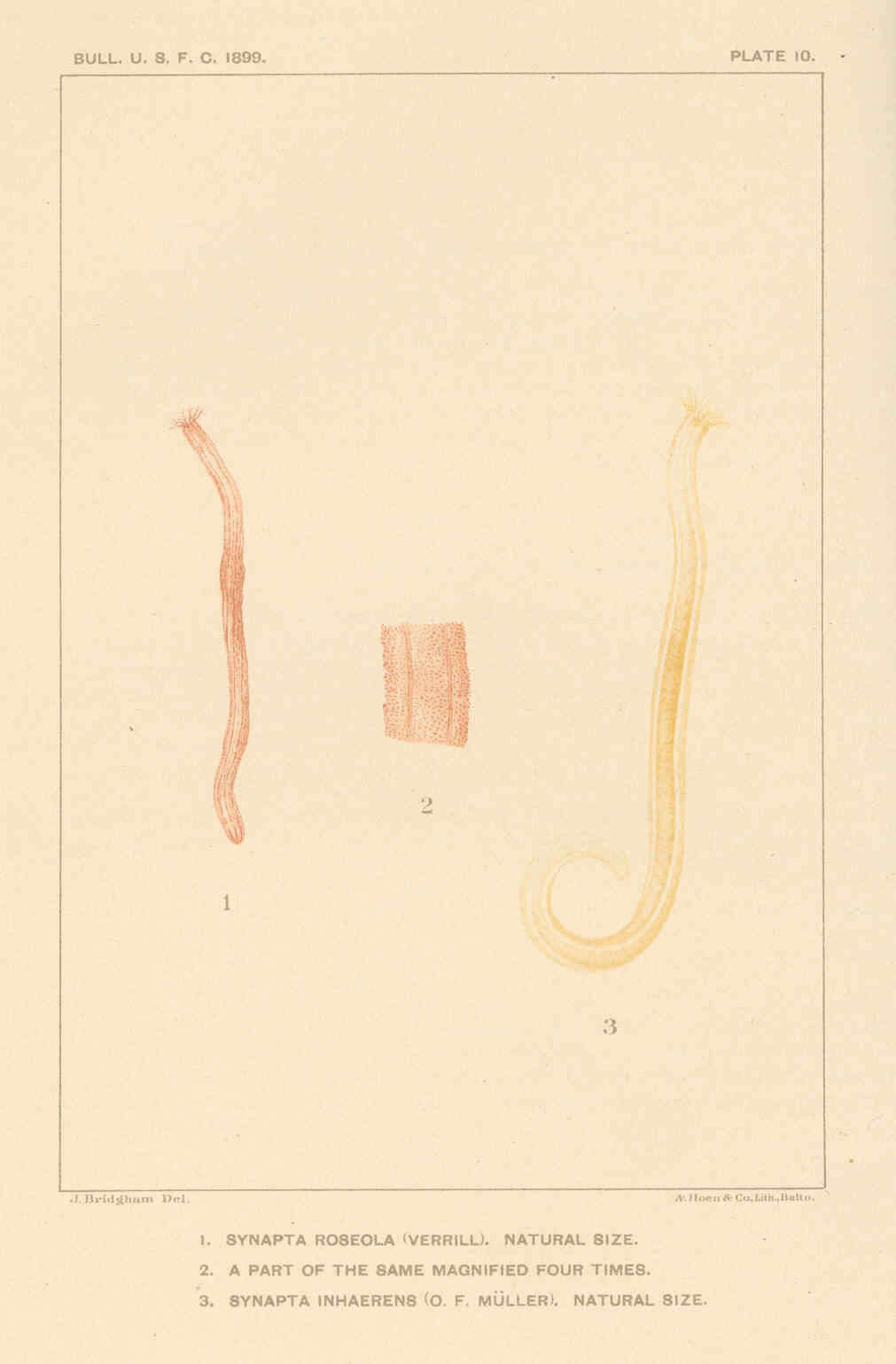
Synapta roseola (Verrill)...(1); Part of the same.... (2); Synapta inhaerens (O. F. Muller)....
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.
-
All Biocode files are based on field identifications to the best of the researcher’s ability at the time.